In the field of cancer diagnostics, precision is paramount. This is where Veracyte’s Afirma Genomic Sequencing Classifier® (GSC) steps in, leveraging the power of whole-transcriptome derived analysis to provide actionable insights into thyroid nodule tumor management. But what exactly is the transcriptome, and how does the Afirma GSC leverage it to help providers and their patients?
Simply put, the word “transcriptome” is the combination of the words “transcript” and “genome”. To fully understand what the transcriptome is and the role that it plays, we need to start at the DNA and end at the protein. This flow of genetic information from DNA to RNA, and then finally to the protein, is referred to as “the central dogma.”
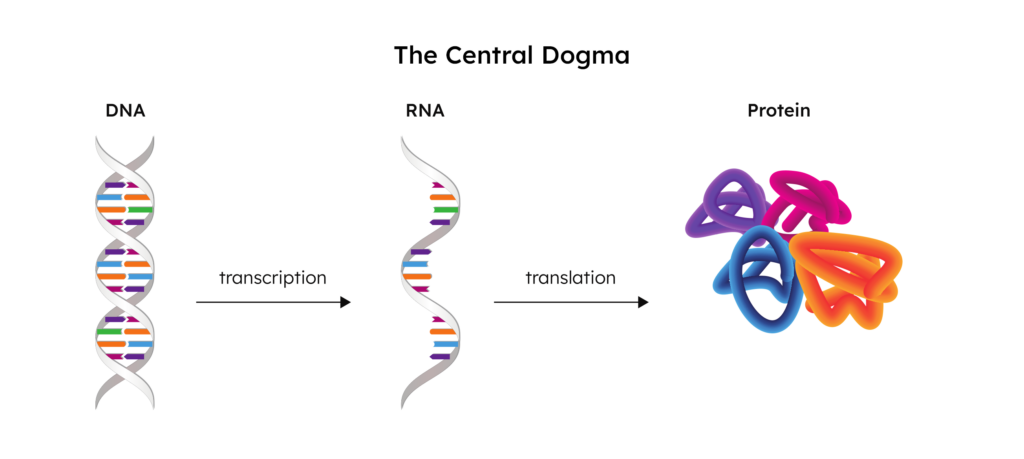
Transcription
The human genome is made up of DNA, the blueprint used to create all of the cells in an organism. When a gene from this DNA is activated (“expressed”), a portion of the DNA is unzipped by RNA polymerase and an RNA copy is created.
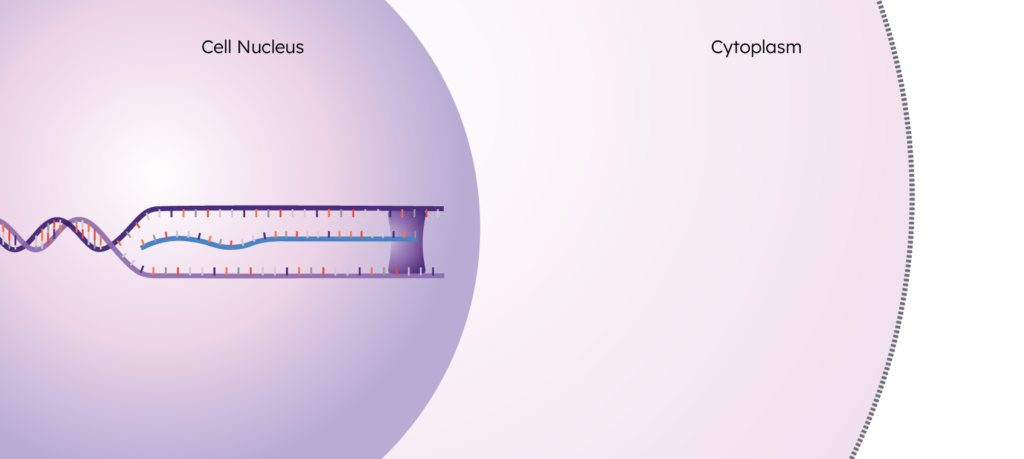
These RNA copies are called “transcripts”, and this process of creating them is called “transcription”. These transcripts exit the nucleus and enter the cytoplasm to eventually undergo translation, resulting in the final step of the central dogma: the protein.
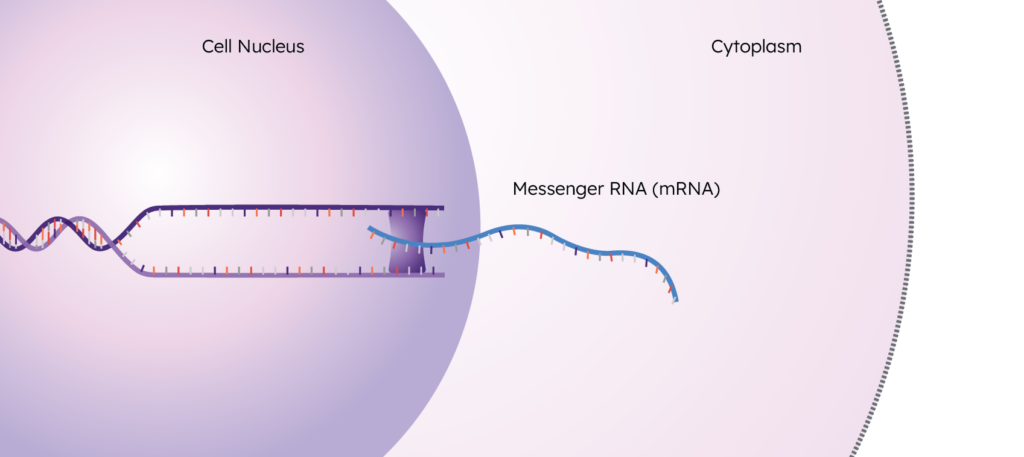
The collection of all of these transcripts in a sample is the transcriptome.
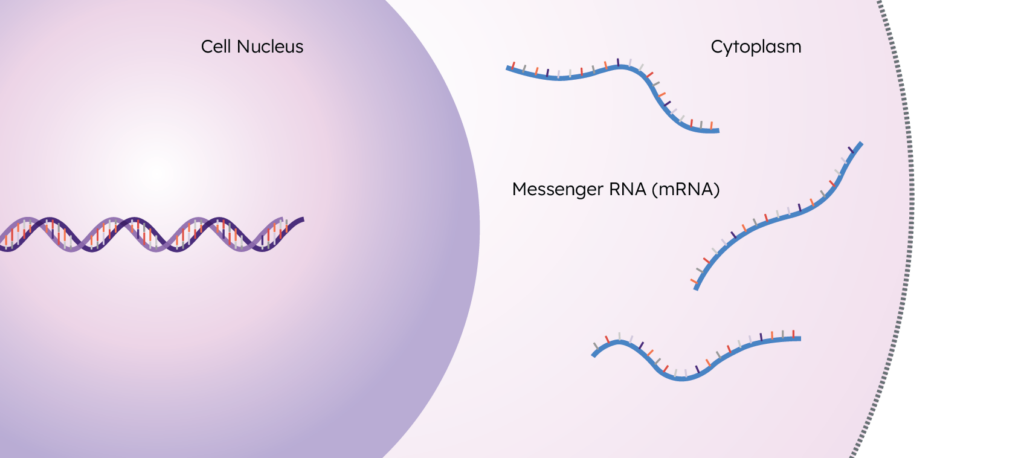
What the Transcriptome Tells Us
As the functional output of the genome, the transcriptome can provide valuable information that the genome can’t. As mentioned earlier, in order for a transcript to exist, a gene must get activated – this is called “gene expression”. By looking at which transcripts are present in a cell, we can see which genes are actually getting expressed – and by looking at how many transcripts are present, we get a quantitative measure of how expressed they are.
This is important, because the presence of a genetic variant does not guarantee its expression. Heart and brain cells feature the same exact DNA, for instance, and yet are entirely different in appearance and function due to mRNA expression. Also, the presence of an alteration may or may not be relevant: some mutations are cancer drivers, some are low-risk, and some are not yet understood.
Assessing the genome provides valuable insight into what alterations are present in a tumor, but in order to understand how the tumor behaves (or how the genome behaves), you have to assess the transcriptome. This cannot be accomplished by looking at DNA alone.
Mitochondrial Transcripts & Loss of Heterozygosity
The genome also misses critical information provided by mitochondrial transcripts due to the nuclear genome being separate from the mitochondrial genome. With whole-transcriptome derived analysis, Afirma GSC is able to leverage both mitochondrial transcripts and chromosomal loss of heterozygosity to classify oncocytic (Hürthle) cell lesions with high sensitivity.1,2
DNA analysis provides valuable information, but it’s not always clinically actionable for these reasons. Since the transcriptome is further along the central dogma and closer to the end product (the protein), it can provide a better picture of what’s actually going on within a cell.
Conclusion
Cancer development is incredibly complex, and often is not caused by a single mutation. This is why Afirma GSC uses whole-transcriptome derived analysis: it better allows us to capture the full complexity of cancer and generate a wealth of valuable information. This information not only serves as the foundation for Afirma GSC, but also creates the potential for novel research. If you’re interested in conducting research of your own or would like to learn more about Afirma GSC, fill out the form below and a genomic specialist will be in touch with you shortly.
References: